Impact Proteomics strives to see what the immune system sees with our advanced immune-profiling technology, IP-to-MS. This immunoassay enables antigen identification and validation, as well as novel antigen discovery. Applications of IP-to-MS include target identification, clinical trial patient stratification, diagnostic development, and immune-related disease research. Our manuscript describing IP-to-MS is now available at the Journal of Proteome Research.
Most traditional immunoassays have a shared shortcoming in that they are targeted and, therefore, biased. Identification of novel antigens has traditionally been a laborious and technically-demanding process. As such, many autoimmune diseases lack detailed molecular characterization and do not have identifiable autoantigen targets. IP-to-MS promises to set a new paradigm for identification of novel autoantigens in autoimmune diseases and of patient-specific cancer antigens.
Importantly, IP-to-MS also offers promise in the realm of precision medicine. While some patients with a given autoimmune disease have a given autoantibody, other patients with that same disease may have a different autoantibody. IP-to-MS can identify these differences in a single, streamlined assay, regardless of whether these specific autoantibodies have previously been characterized.
The IP-to-MS Approach
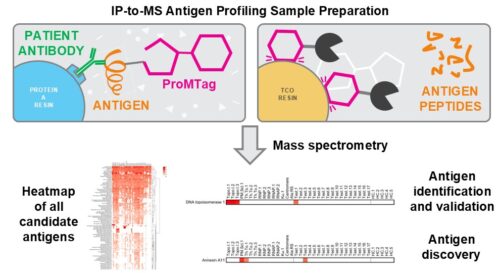
IP-to-MS provides a solution to the overwhelming background noise seen in traditional immunoprecipitation (IP) experiments that utilize mass spectrometry (MS). In IP-to-MS, we harness the power of our reversible protein tag, ProMTag, to separate low abundance immunoprecipitated proteins from highly abundant immunoglobulins under stringent conditions. This allows us to catalogue the antigens associated with a particular disease in a patient-specific manner, using only a small amount of patient serum or plasma.
IP-to-MS begins with binding patient antibodies to protein A resin. At the same time, a pool of target proteins, typically a cell or tissue lysate, is labeled with ProMTag. These are then combined, allowing for ProMTagged antigen proteins to bind to the patient antibodies. After washing away all unbound proteins and contaminants, the protein A resin is stripped, resulting in a pool of ProMTagged antigens and unlabeled antibodies. This is added to TCO resin, to which the ProMTagged proteins will bind. The untagged antibodies are washed away and the antigens are released from the ProMTag, then digested with our MT-Trypsin.
The result of this process is mass-spectrometry ready peptides which require no further cleanup or processing. This streamlined process takes about six hours to complete, and up to 96 samples can be processed at a time.
Learn more about IP-to-MS here.